Implementation of Spatial Transcriptomics
The Laser Microdissection and Transcriptome facilities (Puma, Neurocentre Magendie) wish to develop Spatial Transcriptomics technology.
Indeed, Frédéric Martins, an engineer who recently arrived in the Transcriptome facility, has already carried out this type of experiment in his previous unit. He is therefore looking for interested users.
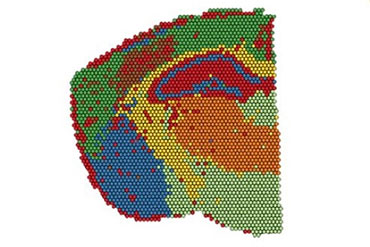
 The goal of Spatial Transcriptomics (Visium technology, 10x Genomics) is to capture mRNAs via their poly-A tail and tag them with a spatial barcode. This barcode then allows them to be analyzed and have their spatial origin identified.
The goal of Spatial Transcriptomics (Visium technology, 10x Genomics) is to capture mRNAs via their poly-A tail and tag them with a spatial barcode. This barcode then allows them to be analyzed and have their spatial origin identified.
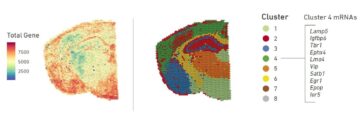
The analysis of the transcriptome is done with a spatial resolution of 50µm and allows us to:
– evaluate the differential expression within different tissue areas (for example between the hippocampus, the thalamus and the hypothalamus)
– know the highest gene expression by area
– generate a list of co-expressed genes
– represent gene-to-gene mapping
This technology requires no investment in terms of equipment and allows us to work on frozen tissue sections (from all organisms) or FFPE (human and mouse) with sizes of 6.5mm x 6.5mm, i.e. the equivalent of a mouse hemisphere. This technology is therefore perfectly suited to the study of the brain.
Contact
PUMA, Neurocentre Magendie
Frédéric Martins
05 57 57 36 79
Last update 16/02/23
