soSMARt: Probing Living Systems at Nanometric Scale and in Depth
Rémi Galland and Jean-Baptiste Sibarita publish in Nature Communications a new super-resolution imaging method, named soSMARt, that enables the investigation of the nanometric organization of proteins at whole cells scale and within 3D multicellular cultures. By combining a light-sheet imaging technique developed in their lab with newly designed imaging devices, this approach paves the way for deeper and more precise exploration of complex 3D biological samples.
Description
Understanding how proteins are organized at the nanometric scale within their natural cellular environment is a key challenge in many areas of biological research. Single Molecule Localization Microscopy (SMLM), a so-called “super-resolution” technique, has for the past two decades pushed the resolution limits of conventional optical microscopy. However, it remains largely restricted to regions close to the coverslip, limiting its ability to image deeper within cells or tissues.
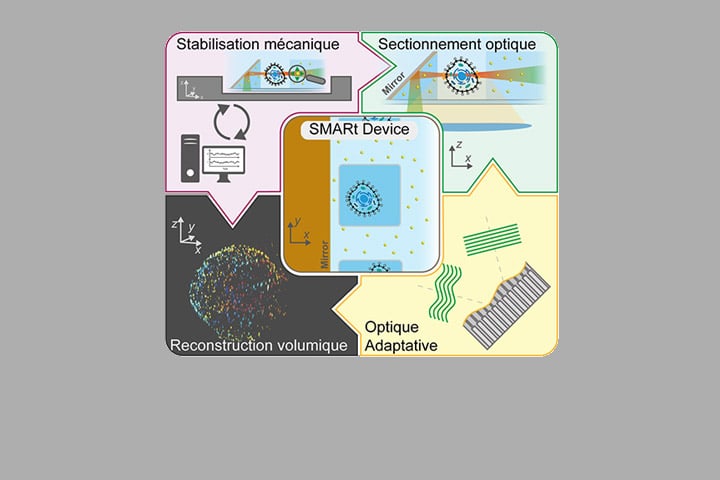
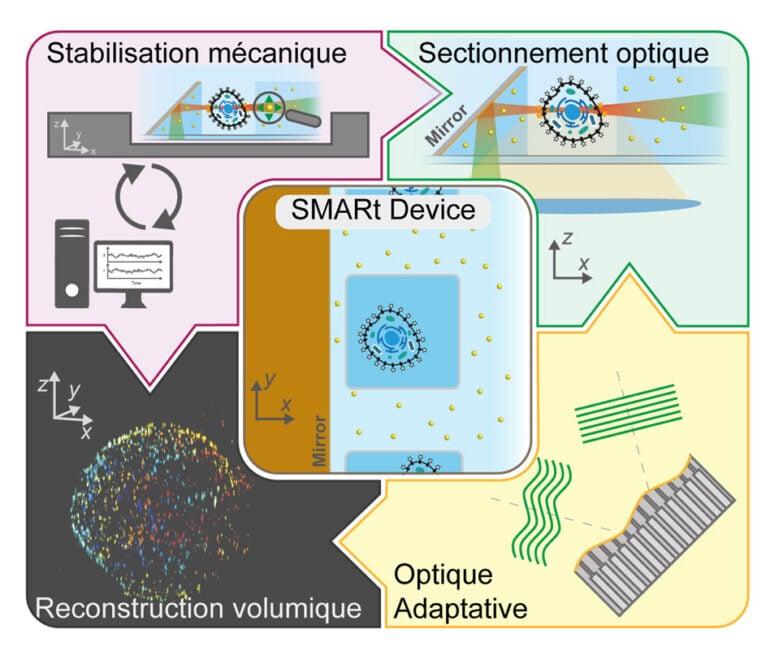
In this context, we developed the soSMARt platform (single-objective Single Molecule Active Registration technique), a method that for the first time combines deep imaging, spatial super-resolution, long-term stability over several hours, and automated acquisition of complete 3D volumes.
The key innovation lies in the use of novel microfabricated devices, called SMARt devices, which integrate:
- 45° inclined mirrors, enabling light-sheet illumination of the sample while maximizing the collection of fluorescence signal (based on the soSPIM technology developed in the lab);
- Photostable fluorescent markers that serve as references for (i) real-time mechanical drift correction, (ii) optical aberration correction via Adaptive Optics (AO), and (iii) nanometric-resolution reconstruction of entire volumes;
A control software (SMARtrack) further ensures active stabilization of the system and fully automates the acquisition of 3D volumes of about 20×20×20 µm³. Thanks to this integrated approach, soSMARt makes it possible to image whole cells or tens of microns deep within multicellular models with spatial resolution as high as 7 nm laterally and 40 nm axially.
We validated this method on various biological systems, ranging from adherent cells, to suspended immune T-cells, and 3D tumor spheroids. This demonstrates the ability of the platform to map the 3D distribution of key proteins in depth, such as PD-1 and CD3 membrane receptors, which are of particular interest in immunotherapy.
Finally, the integration of deep learning-based algorithms enabled us to accelerate the acquisition process by a factor of 10, while improving the quality of the reconstructed super-resolution volumes.
With the soSMARt platform, single-molecule localization microscopy becomes compatible with whole-cell and tissue-like imaging, opening the door to the investigation of the nanometric organization of proteins inside complex 3D samples such as spheroids and organoids, which more accurately mimic physiological conditions found in real organs.
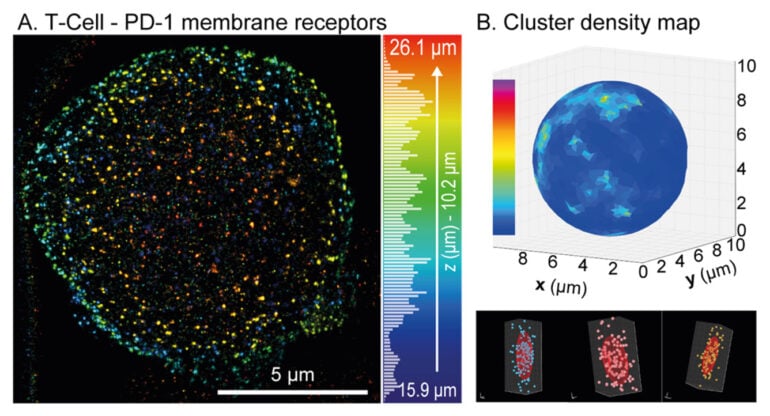
Figure: (A) 3D reconstruction of the PD-1 receptor distribution across the membrane of a T immune cell suspended in a SMARt device, with depth color coding. (B) Projection onto a sphere of the density map of PD-1 receptor clusters (top), with three examples of clusters shown below.
Reference:
Cabillic, H. Forriere, L. Bettarel, C. Butler, A. Neuhaus, I. Idrissi, M.E. Sambrano-Lopez, J. Rossbroich, L-R. Müller, J. Ries, G. Grenci, V. Viasnoff, F. Levet, J-B. Sibarita & R. Galland, “In-depth single molecule localization microscopy using adaptive optics and single objective light-sheet microscopy”, Nature Communications 16, 8362 (2025)
https://doi.org/10.1038/s41467-025-62198-8
Contact
Rémi Galland
Chargé de Recherche CNRS
Institut Interdisciplinaire de Neurosciences (IINS)
CNRS / Université de Bordeaux
Centre Broca Nouvelle Aquitaine
146 rue Léo Saignat
CS 61292 Case 130
F-33076 Bordeaux Cedex (France)
Last update 02/10/25
