Matthieu Lagardère and Olivier Thoumine in Scientific Reports
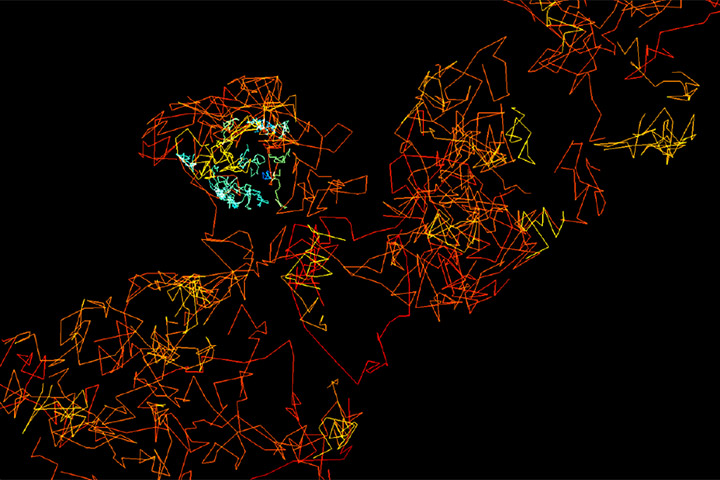
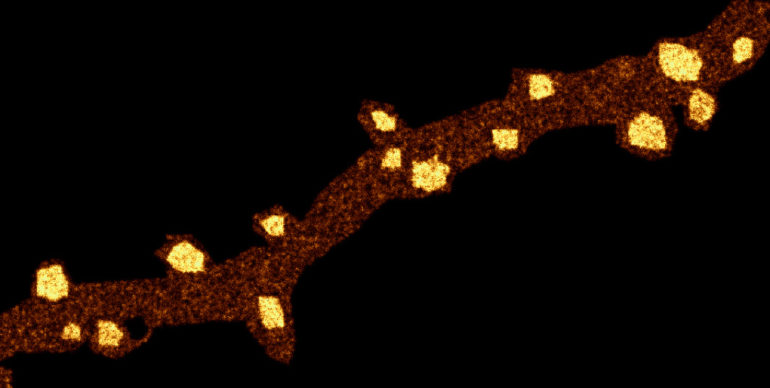
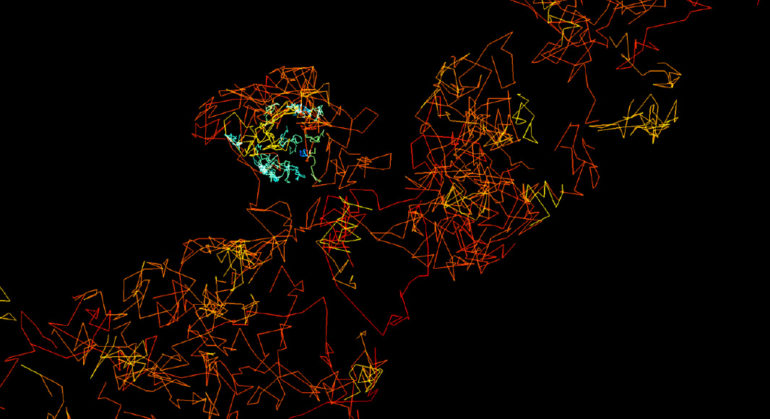
We introduce fast, robust, and user-friendly software called FluoSim that allows for real time simulation of membrane protein dynamics in live-cell imaging and super-resolution modalities. We also show that FluoSim can be used to produce large virtual data sets for training deep neural networks for image reconstruction. This software should thus be of great interest to a wide community specialized in imaging methods applied to cell biology and neuroscience, with the common aim to better understand membrane dynamics and organization in cells. FluoSim is freely available on the website of the publisher at https://www.nature.com/articles/s41598-020-75814-y.
Abstract
Fluorescence live-cell and super-resolution microscopy methods have considerably advanced our understanding of the dynamics and mesoscale organization of macro-molecular complexes that drive cellular functions. However, different imaging techniques can provide quite disparate information about protein motion and organization, owing to their respective experimental ranges and limitations. To address these issues, we present here a robust computer program, called FluoSim, which is an interactive simulator of membrane protein dynamics for live-cell imaging methods including SPT, FRAP, PAF, and FCS, and super-resolution imaging techniques such as PALM, dSTORM, and uPAINT. FluoSim integrates diffusion coefficients, binding rates, and fluorophore photo-physics to calculate in real time the localization and intensity of thousands of independent molecules in 2D cellular geometries, providing simulated data directly comparable to actual experiments. FluoSim was thoroughly validated against experimental data obtained on the canonical neurexin-neuroligin adhesion complex at cell-cell contacts. This unified software allows one to model and predict membrane protein dynamics and localization at the ensemble and single molecule level, so as to reconcile imaging paradigms and quantitatively characterize protein behavior in complex cellular environments.
Reference
Lagardère M 1, Chamma I 1, Bouilhol E 2, Nikolski M 2, Thoumine O 1
FluoSim: simulator of single molecule dynamics for fluorescence live-cell and super-resolution imaging of membrane proteins
Sci Rep 10, 19954 (2020). https://doi.org/10.1038/s41598-020-75814-y
Affiliations
- Univ. Bordeaux, CNRS, Interdisciplinary Institute for Neuroscience, IINS, UMR 5297, F-33000 Bordeaux, France
- Univ. Bordeaux, CNRS, IBGC, UMR 5095, F-33000 Bordeaux, France
Mise à jour: 30/11/20
